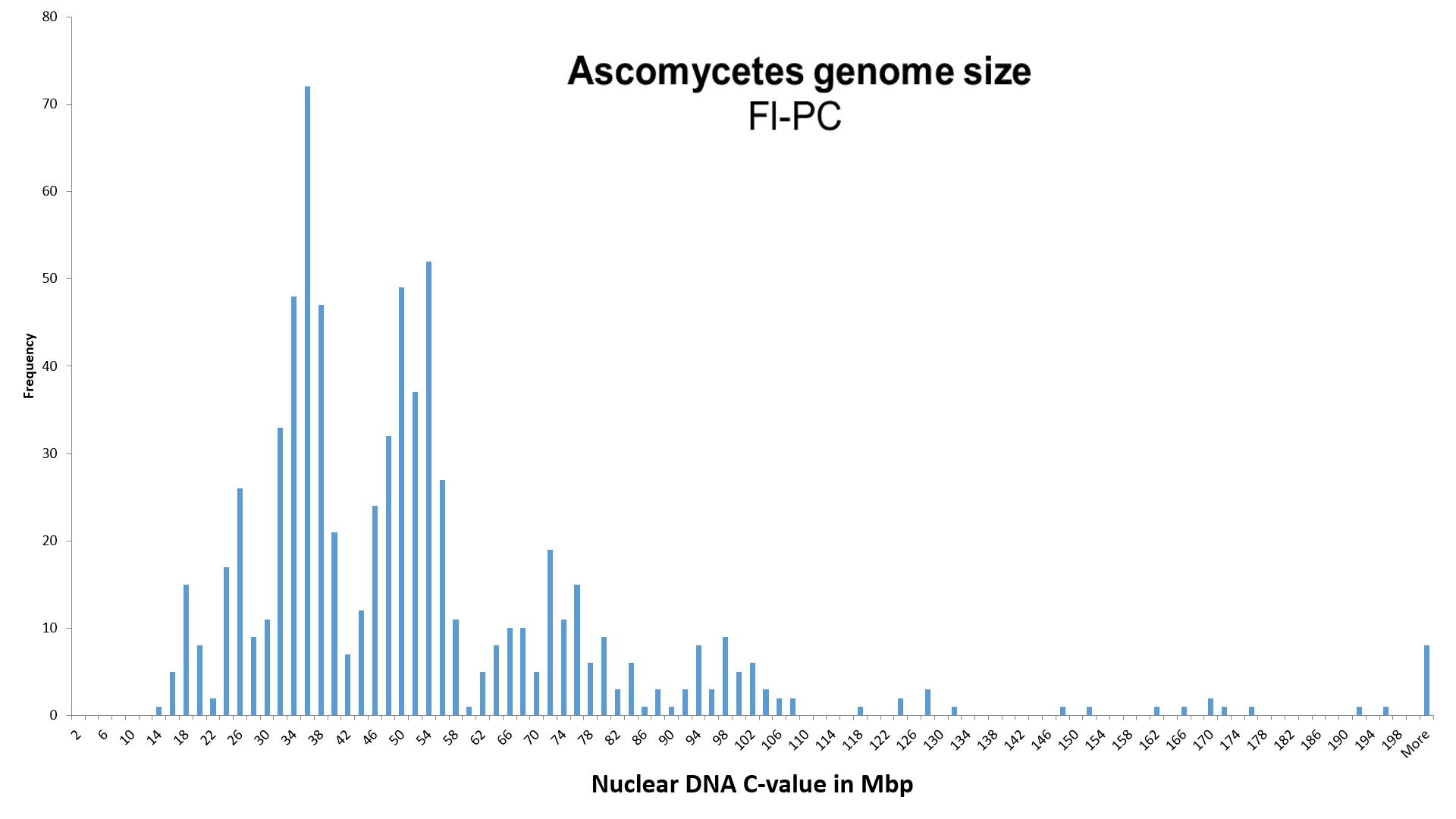
NOTICE! 1) We increased the value of genome size, identified with the Fl-PC method (Photometric Cytometry, stained with Fluorochrome), K = 1.90 times.
By assigning an average value of 25.4 ± 1.3 Mbp (n = 12) of the species Morchella esculenta (= M. conica), used by Weber (1992) as the reference species for measuring the relative genome size of Ascomycota with the Fl-PC method, to the completely sequenced genome size (SC - Sequenced Completely) of 48.21 Mbp of the same species, we obtained correction coefficient K, equal to 1.90 ± 0.10, for the Pl-PC method.
The database contains also other three completely sequenced species (SC), whose genome size was measured before by using the method Fl-PC. Unfortunately, intraspecific variation of the genome size of these species is either not researched (the genome size of Sclerotinia sclerotiorum and Pyronema omphalodes has been measured only once with Fl-PC), or there is involved intraspecific polyploidy (Ascocoryne sarcoides). Taking into account all four completely sequenced species, the corresponding correction coefficient K for the Fl-PC method would be 2.0 ±0.6 (assuming that sequenced A. sarcoides is at ploidy level 2x).
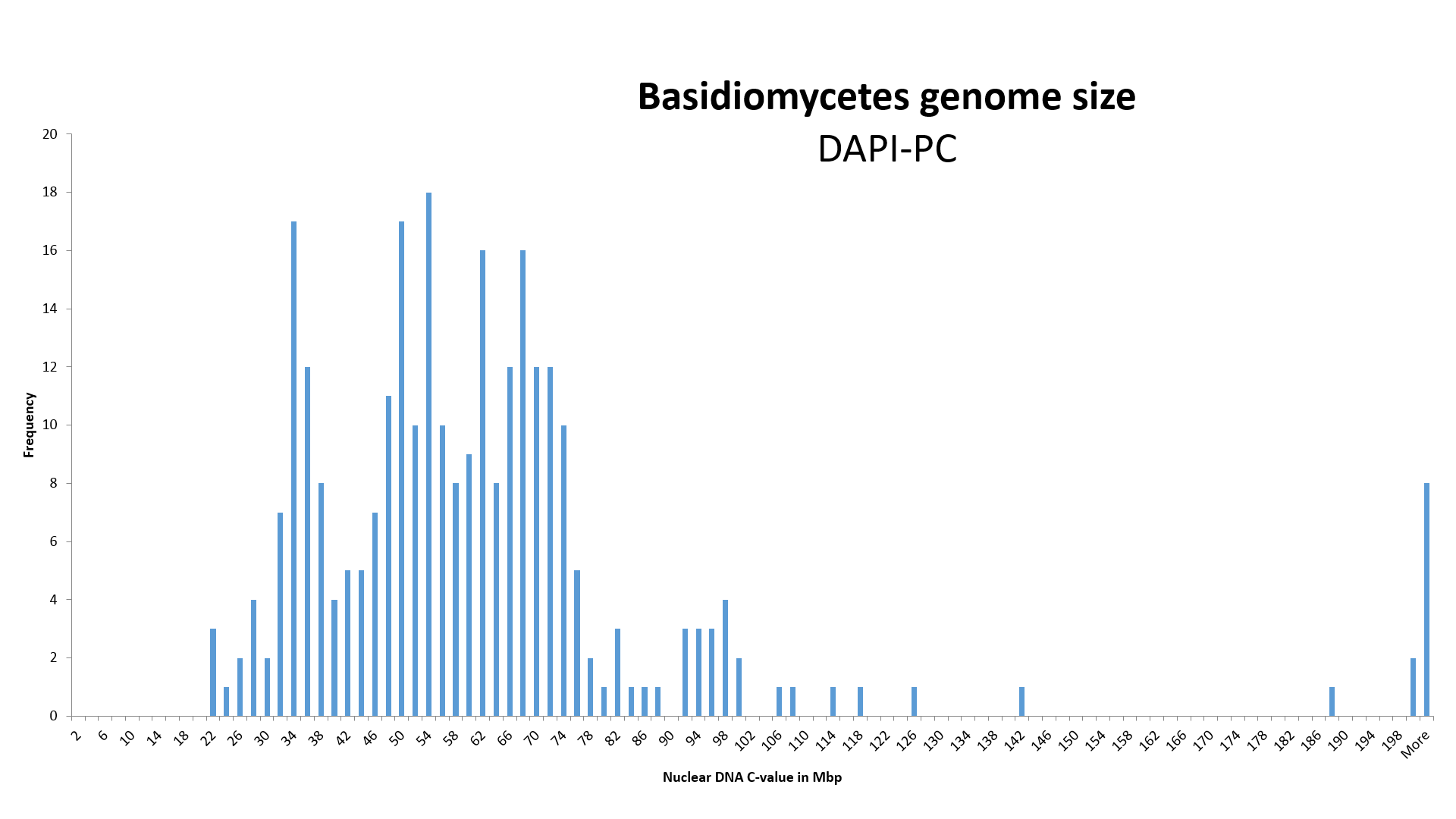
NOTICE! 2) We increased the value of genome size, identified with the DAPI-PC method (Photometric Cytometry, stained with DAPI), K = 1.33 times.
By assigning an average value of 26.3 ± 1.4 Mb (n=7) of the species Pleurotus ostreatus, measured by Wittmann-Meixner (1989) with the DAPI-PC method, to the completely sequenced genome size (SC) of 35.0 ± 0.9 Mbp (n=2) of the same species, we obtained a correction coefficient K, equal to 1.33 ± 0.11, for the genome size obtained with the DAPI-PC method for measuring the relative genome size of Basidiomycota (Wittmann-Meixner, 1989).
Using the CHEF method (Pulsed Field Gel Electrophoresis) the genome size of the same species was measured at 31.4 ± 6.2 Mbp (n=5). In such a case correction coefficient K for the DAPI-PC method would be 1.2.
I measured the genome size of P. ostreatus TAAM126992, using the reference organism Podospora anserina S+ mat 33.77 Mpb (Xu et al, 2007), with DAPI-IC (Image Cytometry, stained with DAPI) and DAPI-FC (Flow Cytometry, stained with DAPI) methods at 32.1 Mbp and 31.9 Mbp respectively (unpublished data). In such a case correction coefficient K for the DAPI-PC method would also be 1.2. If using the reference organism Podospora anserina S+ mat 35.7 Mpb (Espagne et al. et al, 2008), with DAPI-IC (Image Cytometry, stained with DAPI) and DAPI-FC (Flow Cytometry, stained with DAPI) methods at 34.0 Mbp and 33.7 Mbp respectively (unpublished data). In such a case correction coefficient K for the DAPI-PC method would also be 1.3.
We thank Prof. Philippe Silar for the opportunity to use his sequenced strain P. anserina S mat+ as the reference organism.
Bellis Kullman
20150120T1730GMT/UTC+2